Table Of Content

Bisulfite conversion is a process that uses sodium bisulfite to convert all unmethylated cytosines in DNA to uracil. In contrast, methylated cytosines are left unaltered by the sodium bisulfite treatment. After the bisulfite conversion step, a PCR reaction is run to determine the methylation status of CpG sites located between the two primers. During this PCR, the converted uracil bases are read and amplified as thymine, while the methylated cytosines continue to be amplified as cytosine. Thus, cytosines that were methylated in the original DNA will be the only cytosines left in the DNA sequence after amplification. Based on this principle it is possible, by sequencing the bisulfite PCR product, to identify the methylation status of the cytosines in the original DNA sample.
Sign up and select NEB email newsletters targeted to your research
The value of this parameter is less than the actual concentration of oligos in the reaction because it is the concentration of annealing oligos, which in turn depends on the amount of template (including PCR product) in a given cycle. This concentration increases a great deal during a PCR; fortunately PCR seems quite robust for a variety of oligo melting temperatures. Primer-blast tries to find target-specific primers by placing candidate primers on unique template regions that are not similar to other targets. Once the designing of qPCR primers and probes has been done using available tools, insilico validation is to be performed by BLAST (insilico validation) for the confirmation of targeted gene sequences specificity. The algorithm of BLAST carries out sequence- similarity search against several databases with a set of gapped alignments of links to full database records (Raymaekers M et al, 2009). The query coverage and the maximum identity should be 100%.
NCBI
Many of the Swift products you have grown to love are now part of our new complete portfolio, xGen™ NGS. Through this new partnership we are pleased to offer you comprehensive next generation sequencing solutions. Lyophilized primers should be dissolved in a small volume of distilled water or TE to make a concentrated stock solution. Prepare small aliquots of working solutions containing 10 pmol/µl to avoid repeated thawing and freezing.
Primer Design and Fragment Assembly Using NEBuilder HiFi DNA Assembly® or Gibson Assembly®
These primers are then used by the DNA polymerase to build new complementary strands, which will result in the doubling of the number of DNA molecules of the targeted DNA sequence for each PCR cycle. From design to synthesis, quality primers are vital to successful results. Use our online Applied Biosystems™ Primer Designer™ Tool to search for the right PCR/Sanger sequencing primer pair from a database of ~650,000 predesigned primer pairs for resequencing the human exome and human mitochondrial genome. Choose from different amplicon lengths to accommodate various research applications and biological sample types.
Optimised Application Oligos
The millimolar concentration of salt (usually KCl) in the PCR. The following content focuses on demonstrating designing primers for different cloning methods. To follow along, access the sequence of Dsup from this link. With this option on, the program will automatically retrieve the SNP information contained in template (using GenBank accession or GI as template is required) and avoid choosing primers within the SNP regions. The maximum number of PCR targets (amplicons) to be shown when checking specificity for pre-designed primers.
Selecting the best paint primer based on the specifics of your next project will help you achieve great results. Melting temperature (52°C-56°C) The GC results of the sequence gives a fair indication of the primer Tm. However, the difference of the primer should not be less than 2°C. However, other methods would need additional sequences that aid in ligation / recombination or determining the directionality of the insert. There are certain guidelines to follow when designing a primer.

In general, the non-specific targets become less of a concern if their sizes are very large since PCR is much less efficient for larger amplicons. What's more, the latest formulas now feature skincare ingredients, such as niacinamide for oily skin, glycerin and vitamin E for moisture, and antioxidants to address damage from sun and pollutants. Others have a built-in tint or color-correcting properties, so you might not even need makeup once you're done. With that in mind, if you want to give primer a try, we already did all the testing for you. Here are the best primers, according to Glamour editors. Alternatively, you can reverse complement the sequence without copy-pasting the sequence into a new window.
A novel tailed primer nucleic acid test for detection of HPV 16, 18 and 45 DNA at the point of care Scientific Reports - Nature.com
A novel tailed primer nucleic acid test for detection of HPV 16, 18 and 45 DNA at the point of care Scientific Reports.
Posted: Tue, 21 Nov 2023 08:00:00 GMT [source]
Sequence selected for designing forward primer (text highlighted in blue that is pointed by arrowhead). The red box indicates the parameters of the selected sequence like Tm, length of the sequence, and GC%. The reverse primer was designed from the end of the sequence that we have added to APE. This article demonstrates how to design primers (forward and reverse) for different types of cloning methods. While this system is incredibly useful to amplify and precisely quantify a gene of interest, there are several obstacles that can lower primer efficiencies and jeopardize your experiment.
While this system is incredibly useful to amplify and precisely quantify a gene of interest, there are several obstacles that can jeopardize your experiment. Overcoming low primer efficiency is the first step to a successful PCR, but how can you ensure your PCR product is concentrated and pure enough for downstream applications? Zymo Research’s DNA Clean & Concentrator Kits are capable of maximizing DNA concentration and removing contaminants, making your PCR product suitable for even the most sensitive downstream applications. The maximum number of PCR targets (amplicons) to be found on any single sequence in the search database.
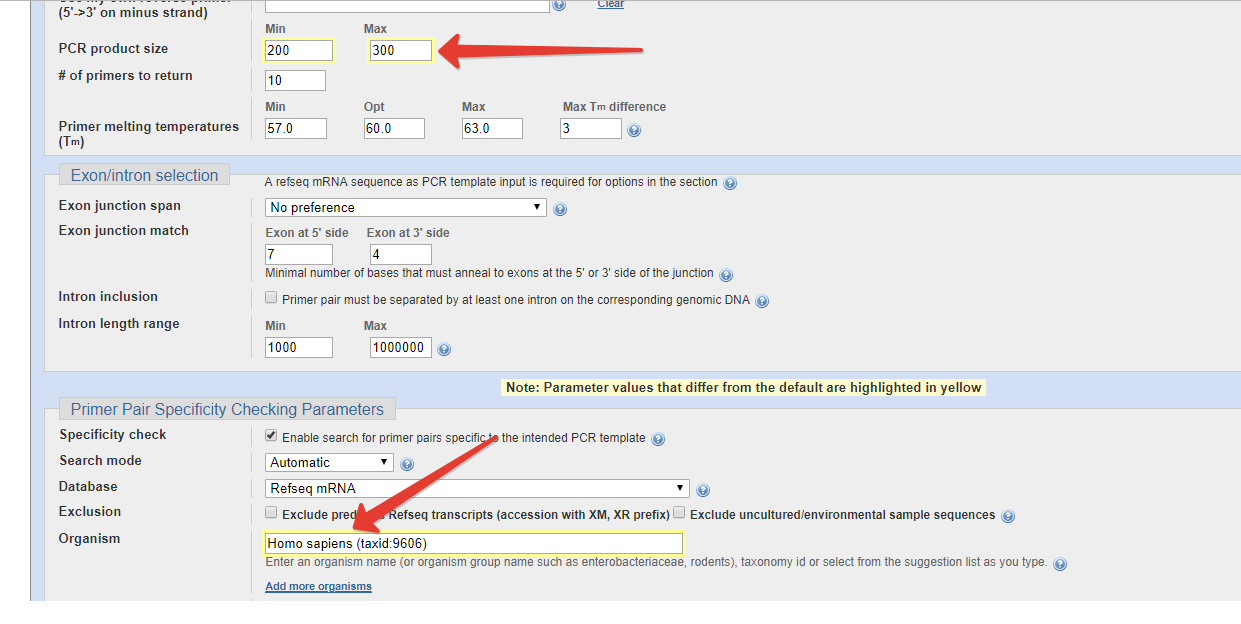
This will limit the primer specificity checking to the specified organism. It is strongly recommended that you always specify the organism if you are amplifying DNA from a specific organism (because searching all organisms will be much slower and off-target priming from other organisms is irrelevant). Click on "Add more organisms" label if you want to restrict to multiple organisms (enter only one organism in each input box).
In these cases, you need to modify the primer designing strategy or the amplification strategy. Illustration depicting the design of forward primer and its involvement in PCR. In case, if you take the complementary sequence and use it for primer designing (laying the sequence in 5’-3’), the reverse primer in the above example now acts as the forward primer. Zymo Research’s DNA Clean & Concentrator Kits were designed to facilitate the removal of salts, polymerases, and endonucleases from your PCR product, leaving you with purified, highly concentrated DNA.
The resulting DNA is suitable for even the most sensitive downstream applications including sequencing, cloning, ligation, and endonuclease digestion. Discover which PCR purification kit best suits your research goals today. The structure of the primer should be relatively simple and contain no internal secondary structure to avoid internal folding. One also needs to avoid primer-primer annealing which creates primer dimers and disrupts the amplification process. When designing, if unsure about what nucleotide to put at a certain position within the primer, one can include more than one nucleotide at that position termed a mixed site.
The following table gives the comparison of primers designed for various cloning methods. Illustration depicting the design of reverse primer and its involvement in PCR. The sequence covered in blue open rectangle is used for reverse primer. Blue ribbon indicates primer sequence selected for reverse primer design. Reverse complementation contains two steps (Complementation and changing the direction) were shown.
This method demands the use of a vector assembly (plasmid) into a single construct with one or multiple DNA fragments. The PCR primers overlap to form restriction sites with adjacent DNA fragments and are designed, however, Type 2S enzyme, along with DNA ligases of the fragments for a directional assembly. Likewise, this method exploits the use of type 2 class of restriction sites, i.e. cut outside of their restriction sites through non-palindromic sticky end overhangs. This method exploits multiple fragments of DNA by using a combination of overhang sequences on their insert fragments for easy annealing with the adjacent ones.
To reverse complement, copy the sequence (ctrl+c or cmd+c) and past the sequence (ctrl+v or cmd+v) in new window (Fig 4.1). There is a button in the header part of the APE to reverse complement the sequence. Select the primer sequence (if it is not selected already) and click on the button (Fig 4.2).
A higher E value should be used if you want more stringent specificity checking (i.e., to identify targets that have more mismatches to the primers, in addition to the perfectly matched targets). On the other hand, a lower E value is recommended if you are only interested in perfect or nearly perfect matches as this will significatly shorten the search time. Enter the position ranges if you want the primers to be located on the specific sites.
No comments:
Post a Comment